From preclinical hope to clinical proof, without the guesswork.
Scienta develops the first foundation model predicting human immune response, bringing clarity to translational research in immunology & inflammation.
At Scienta, we developed a predictive layer that turns early biological data into reliable clinical insight.





From target ID to clinical plan, already mapped out.


Target and mechanism prioritization
Targets and mechanisms are ranked by predicted probability of clinical success using integrated multi-omic data, disease biology and early experimental signals, giving teams a clear view of which programs deserve priority before major resources are committed.


Preclinical-to-human translation
Human efficacy is predicted from preclinical and target-level data by simulating drug impact on immune pathways and translating these effects into expected clinical outcomes, enabling objective candidate comparison and early go / no-go decisions ahead of IND.


Patient stratification and trial optimization
Patient-level response is predicted from baseline and post-treatment data to identify biomarkers and support trial enrichment strategies, improving signal detection and accelerating decision-making in Phase II–III development.
ACCURACY OF CLINICAL TRIAL SUCCESS PREDICTIONS
immuno-inflammatory diseases covered
tissues integrated across datasets
PATIENT AND BIOSAMPLE PROFILES ACROSS diseases
Powered by tech. Guided by people.
EVA
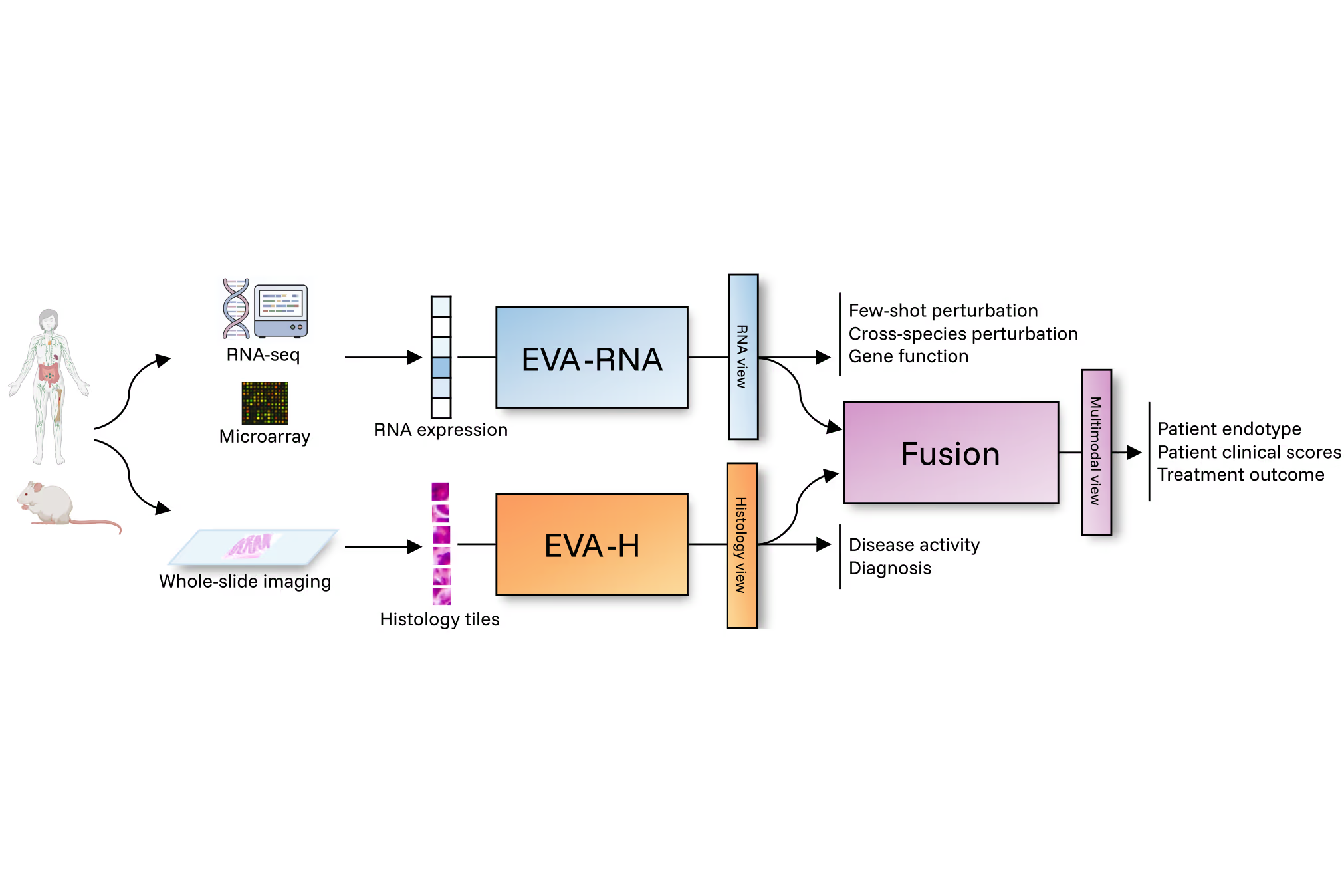
EVA is Scienta's foundation model, designed specifically for immuno-inflammatory diseases. It learns biology across conditions, revealing the shared mechanisms driving these pathologies.
Expert guidance

Each project is supported by immunologists and data scientists to translate predictions into concrete development decisions, from target selection to trial strategy.
ImmunAtlas

ImmunAtlas is Scienta Lab's proprietary dataset used to train EVA. It combines multi-omic, clinical and histopathology data from more than 96,000 patient and biosample profiles and continues to grow as new datasets are integrated.

Hear it from them
News and publications
Do you have any questions?
Can EVA generate predictions for first-in-class assets and complex modalities such as bispecifics and combination therapies?
Yes. EVA is designed to generalize beyond known drugs and mechanisms.
It can generate predictions for novel targets and first-in-class assets, as well as model the combined impact of multiple targets or therapeutic mechanisms by simulating how they jointly perturb immune networks and influence patient-level outcomes.
How much data is required to obtain a reliable prediction?
At the discovery stage, EVA can generate predictions without any asset-specific data by relying on immune biology learned across diseases, tissues and patient populations.
For a specific asset in preclinical development, relevant experimental data is required. EVA is built to operate under realistic R&D constraints, even with limited datasets, and prediction confidence increases as additional data becomes available.
What data should I provide and will it remain confidential?
EVA can integrate a wide range of data types, including transcriptomics (bulk or single-cell RNA-seq), proteomics, clinical scores, histopathology, target information and preclinical readouts.
Your proprietary data remains fully confidential and is used only to fine-tune a private instance of the model for your project. It is never shared, reused or incorporated into the global training of EVA.
What makes EVA different from generic AI models or standard dataset analysis?
EVA is a foundation model purpose-built for immunology and inflammation.
It is trained on immune-specific biological and clinical data and designed to capture the unique complexity of immune-mediated diseases.
Beyond single-dataset analysis, EVA applies transfer learning across diseases, tissues and patient populations, allowing robust, biologically grounded predictions even from limited new data; something generic AI models and standard analytics cannot provide.